Using Supervised Convex Clustering for Two Dimensional Clusters
Title: Supervised Convex ClusteringAuthors & Year: Minjie Wang, Tianyi Yao, and Genevera I. Allen (2023)Journal: Biometrics [DOI: 10.1111/biom.13860]Review Prepared by Edmond Sanou Clustering is a common way for computers to group together similar things when we don’t already know the categories. The problem is that the groups it finds are sometimes hard to make sense of. In real life, though, we often have extra pieces of information, like doctors’ opinions or other clues, that, while imperfect, can help guide the grouping. The authors Wang, Yao, and Allen. created a new method called supervised convex clustering (SCC), which combines both the raw data and these extra guiding clues. This makes the discovered groups easier to interpret and more meaningful scientifically. They also adapted the method so it can handle different kinds of extra information and even look for patterns in two dimensions at once (biclusters). When they tested SCC, including on…
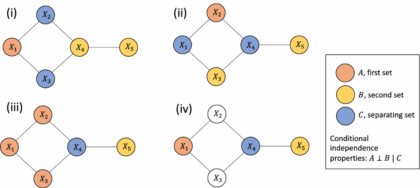
How can Gaussian graphical Networks Models help in exploring Ovarian cancer data?
Article: Gaussian graphical models with applications to omics analysesAuthors and Year: Katherine H. Shutta, Roberta De Vito, Denise M. Scholtens, Raji Balasubramanian 2022Journal: Statistics in Medicine Review Prepared by: Sanou Edmond, Postdoc in BiostatisticsNuclear Safety and Radiation Protection Authority (ASNR) As scientists collect more detailed biological data, they use networks to understand how molecules in the body interact and how these interactions relate to disease. This type of data, known as omics, includes information about genes (genomics), proteins (proteomics), and other molecules. These networks can help find genes linked to illness and even suggest possible treatment options. Statisticians help by using tools that highlight which molecules are directly connected. In their tutorial “Gaussian Graphical Models with Applications to Omics Analyses,” Shutta et al. recommend using a method called Gaussian Graphical Models (GGMs) to study these connections. GGMs help draw simple, clear maps of how molecules relate to each other. The authors…